Neural Network#
Neural Networks are a machine learning model that learns the optimal parameters to approximate complex functions.
GitHub Repo: gavinkhung/neural-network
Import the libraries
import numpy as np
%matplotlib ipympl
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from celluloid import Camera
import scienceplots
from IPython.display import Image
np.random.seed(0)
plt.style.use(["science", "no-latex"])
Training Dataset#
The neural network will learn the parameters to fit a Hyperbolic Paraboloid.
\( \begin{align*} z &= \frac{y^2}{b^2} - \frac{x^2}{a^2} \end{align*} \)
def generate_function(dims):
a = 1
b = 1
# Hyperbolic Paraboloid
x = np.linspace(-1, 1, dims)
y = np.linspace(-1, 1, dims)
X, Y = np.meshgrid(x, y)
Z = (Y**2 / b**2) - (X**2 / a**2)
X_t = X.flatten()
Y_t = Y.flatten()
Z_t = Z.flatten()
X_t = X_t.reshape((len(X_t), 1))
Y_t = Y_t.reshape((len(Y_t), 1))
Z_t = Z_t.reshape((len(Z_t), 1))
features = np.stack((X_t, Y_t), axis=1)
labels = Z_t.reshape((len(Z_t), 1, 1))
return X, Y, Z, features, labels
dims = 12
X, Y, Z, features, labels = generate_function(dims)
# Visualize the Hyperbolic Paraboloid
fig = plt.figure()
ax = fig.add_subplot(111, projection="3d")
ax.plot_surface(X, Y, Z, color="red", alpha=0.5)
<mpl_toolkits.mplot3d.art3d.Poly3DCollection at 0xffff6c1f8530>
Loss Function#
The loss function is needed to evaluate the performance of the model and to update the weights accordingly. The optimizaton process of the neural network training will find the weights and biases to minimie the loss.
Mean Squared Error#
Quadratic loss functions, like mean squared error, are used for regression tasks, like this example.
\( J = \frac{1}{n} \sum_{i=1}^{n}(y_{i}-\hat{y})^2 \)
def mse(y_true, y_pred):
return np.mean(np.power(y_true - y_pred, 2))
Mean Squared Error Gradient#
In order to perform backpropagation to update the weights, we need to calculate the gradient of the loss function.
\( \begin{align*} J^{\prime} &= \frac{\partial}{\partial \hat{y}} [ \frac{1}{n} \sum_{i=1}^{n}(y_{i}-\hat{y})^2 ] \\ &= \frac{1}{n} \sum_{i=1}^{n}\frac{\partial}{\partial \hat{y}} [ (y_{i}-\hat{y})^2 ] \\ &= \frac{2}{n} \sum_{i=1}^{n} (y_{i}-\hat{y}) \frac{\partial}{\partial \hat{y}}[y_{i}-\hat{y}] \\ &= \frac{2}{n} \sum_{i=1}^{n} (y_{i}-\hat{y}) (-1) \\ &= \frac{2}{n} \sum_{i=1}^{n}(\hat{y}-y_{i}) \end{align*} \)
def mse_prime(y_true, y_pred):
return 2 * (y_pred - y_true) / np.size(y_true)
Neural Network Layer#
We will implement a feedforward neural network, which contains many layers. Each layer at a very high applies a linear transformation to the input data to create the output data, which is often in a different dimension than the input data. Then, all of the values in the output are run through a function, known as an activation function.
Recall from Linear Algebra that an input column vector \(x\) in \(\mathbb{R}^n\) can be transformed into another dimension \(\mathbb{R}^m\) by multipying it with a matrix of size \(m \times n\). Finally we can add a bias term to this linear transformation to shift the transformed data up or down.
As a result, each layer stores a weight matrix and a bias vector to apply the linear transformation.
from abc import ABC, abstractmethod
class Layer(ABC):
def __init__(self):
self.input = None
self.output = None
self.weights = None
self.bias = None
@abstractmethod
def forward(self, input):
pass
@abstractmethod
def backward(self, output_gradient, optimizer):
pass
Forward Propagation#
This the process of transforming our input data into the predictions from our feedforward neural network. The input data is passed through every layer of the network. Below is a visualization of the 3 layer network we will implement.
Each neuron in a layer is a weighted sum of its inputs plus a bias term. Finally, an activation function is applied on each neuron in the layer. We will have a class to represent a fully-connected layer and another class to represent an activation function.
The computation for each neuron in a layer being a weighted sum of the products between the inputs and the layer’s weights plus a bias term is the same as a matrix multiplication between the weight matrix and the input data added with a bias vector.
Thus, the foward propagation of a fully connected layer can be written as:
\( \begin{align*} Z &= W X + B \\ A &= g(Z) \end{align*} \)
Where:
\(W\) is the weight matrix for the layer
\(X\) is the input data
\(B\) is a bias vector
\(g\) is the activation function
Our Network’s Forward Propagation#
We will implement a 3-layer fully-connected neural network with a Tanh activation function after each layer. These transformations can be written as these matrix multiplications:
\( \begin{align*} Z_1 &= W_1 X + B_1 \\ A_1 &= tanh(Z_1) \\ \\ Z_2 &= W_2 A_1 + B_2 \\ A_2 &= tanh(Z_2) \\ \\ Z_3 &= W_3 A_2 + B_3 \\ \hat{y} &= A_3 = tanh(Z_3) \end{align*} \)
Backpropagation#
Backpropagation is the process of updating all of the weights and biases in a neural network using the chain rule and gradient descent.
The following equations use \(J\) as our loss function. In this Notebook, we use the mean squared error loss function. Click here for the mean squared error loss function.
Our goal is to apply gradient descent on the weights and bias using the following equations:
\( \begin{align*} W_1 &= W_1 - lr * \frac{\partial J}{\partial W_1} \\ B_1 &= B_1 - lr * \frac{\partial J}{\partial B_1} \\ W_2 &= W_2 - lr * \frac{\partial J}{\partial W_2} \\ B_2 &= B_2 - lr * \frac{\partial J}{\partial B_2} \\ W_3 &= W_3 - lr * \frac{\partial J}{\partial W_3} \\ B_3 &= B_3 - lr * \frac{\partial J}{\partial B_3} \\ \end{align*} \)
We need to use the chain rule to get the values for \(\frac{\partial J}{\partial W_1}\), \(\frac{\partial J}{\partial B_1}\), \(\frac{\partial J}{\partial W_2}\), \(\frac{\partial J}{\partial B_2}\), \(\frac{\partial J}{\partial W_3}\), and \(\frac{\partial J}{\partial B_3}\).
Chain Rule for Backpropagation#
Let’s derive a general formula to update the weight matrix \(W_i\) and bias vector \(B_i\) of a single fully-connected layer.
To apply gradient descent on the 3rd layer, for example, we need to find the loss with respect to \(W_3\) and \(B_3\), which are \(\frac{\partial J}{\partial W_3}\), and \(\frac{\partial J}{\partial B_3}\).
The loss function \(J\) is in terms of \(A_3\), which is the final output/activation of the last layer. Then, \(A_3\) is in terms of \(Z_3\). Lastly, \(Z_3\) is in terms of \(W_3\), \(B_3\), and \(A_2\), which is the activation from the second to last layer. Let’s get the loss with respect to \(W_3\) and \(B_3\).
We can represent these matrix operations as the following composite functions: \(J(A_3)\), \(A_3(Z_3)\), and \(Z_3(W_3, B_3, A_2)\)
Chain Rule For Weight Matrix W#
Use the chain rule to derive \(\frac{\partial J}{\partial W_3}\)
\( \begin{align*} \frac{\partial J}{\partial W_3} &= \frac{\partial}{\partial W_3}[J(A_3(Z_3(W_3, B_3, A_2)))] \\ &= \frac{\partial J}{\partial A_3} \frac{\partial}{\partial W_3}[A_3(Z_3(W_3, B_3, A_2))] \\ &= \frac{\partial J}{\partial A_3} \frac{\partial A_3}{\partial Z_3} \frac{\partial}{\partial W_3}[Z_3(W_3, B_3, A_2))] \\ &= \frac{\partial J}{\partial A_3} \frac{\partial A_3}{\partial Z_3} \frac{\partial Z_3}{\partial W_3} \\ \end{align*} \)
Chain Rule For Bias Vector B#
Use the chain rule to derive \(\frac{\partial J}{\partial B_3}\).
\( \begin{align*} \frac{\partial J}{\partial B_3} &= \frac{\partial}{\partial B_3}[J(A_3(Z_3(W_3, B_3, A_2)))] \\ &= \frac{\partial J}{\partial A_3} \frac{\partial}{\partial B_3}[A_3(Z_3(W_3, B_3, A_2))] \\ &= \frac{\partial J}{\partial A_3} \frac{\partial A_3}{\partial B_3} \frac{\partial}{\partial B_3}[Z_3(W_3, B_3, A_2))] \\ &= \frac{\partial J}{\partial A_3} \frac{\partial A_3}{\partial Z_3} \frac{\partial Z_3}{\partial B_3} \\ \end{align*} \)
Backpropagation for Weight Matrix W#
\( \begin{align*} \frac{\partial J}{\partial W_3} = \frac{\partial J}{\partial A_3} \frac{\partial A_3}{\partial Z_3} \frac{\partial Z_3}{\partial W_3} \end{align*} \)
Let’s break each component down:
\(\frac{\partial J}{\partial A_3}\) is the gradient of the loss function, which is the gradient of the mean squared error function. Click here for the derivation of the loss function gradient. In the general case for any layer, this is the gradient from the next layer (idx+1).
\(\frac{\partial A_3}{\partial Z_3} = \frac{\partial}{\partial Z_3}[tanh(Z_3)] \) is the gradient of the activation function. Click here for the derivation of the activation function gradient.
\(\frac{\partial Z_3}{\partial W_3} = \frac{\partial}{\partial W_3}[W_3 A_2 + B_3] = A_2\) is the original input to the layer, which is the output of the previous layer (idx-1).
As a result, to the gradient of the weight matrix of a fully-connected layer is the matrix multiplication products of the following:
The gradient from the next layer (idx+1)
The gradient of the activation function
The input from the previous layer (idx-1)
Backpropagation for Bias Vector B#
\( \begin{align*} \frac{\partial J}{\partial B_3} = \frac{\partial J}{\partial A_3} \frac{\partial A_3}{\partial Z_3} \frac{\partial Z_3}{\partial B_3} \end{align*} \)
Let’s break each component down:
\(\frac{\partial J}{\partial A_3}\) is the gradient of the loss function, which is the gradient of the mean squared error function. Click here for the derivation of the loss function gradient. In the general case for any layer, this is the gradient from the next layer (idx+1).
\(\frac{\partial A_3}{\partial Z_3}\) is the gradient of the activation function. Click here for the derivation of the activation function gradient.
\(\frac{\partial Z_3}{\partial B_3} = \frac{\partial}{\partial B_3}[W_3 A_2 + B_3] = 1\) is 1, we can ignore this in the gradient computation.
As a result, to the gradient of the bias vector of a fully-connected layer is the matrix multiplication products of the following:
The gradient from the next layer (idx+1)
The gradient of the activation function
For more information, I recommend the follow resources:
Neural Network from Scratch. I also watched this video to help write this Notebook.
Dense Layers#
A dense layer is a fully connected layer. Let’s use the equations derived in the forward and backwards propagation sections above to implement the forward() and backward() methods of our dense layer class.
class Dense(Layer):
def __init__(self, input_neurons, output_neurons):
# Random Values from Normal Distribution
# self.weights = np.random.randn(output_neurons, input_neurons)
# self.bias = np.random.randn(output_neurons, 1)
# All Zeros
# self.weights = np.zeros((output_neurons, input_neurons))
# self.bias = np.zeros((output_neurons, 1))
# Xavier Initialization Uniform Distribution
limit = np.sqrt(6 / (input_neurons + output_neurons))
self.weights = np.random.uniform(
-limit, limit, size=(output_neurons, input_neurons)
)
self.bias = np.zeros((output_neurons, 1))
def forward(self, input):
self.input = input
return np.matmul(self.weights, self.input) + self.bias
def backward(self, output_gradient, optimizer):
# Calculate gradients
weights_gradient = np.matmul(output_gradient, self.input.T)
input_gradient = np.dot(self.weights.T, output_gradient)
# Update weights and biases
self.weights, self.bias = optimizer.backward(
self.weights, weights_gradient, self.bias, output_gradient
)
return input_gradient
Activation Function#
Activation functions are applied after the matrix multiplication to introduce nonlinearity into our neural networks. Choosing the correct activation function is essential for the neural network to learn general patterns of the training data. Most notably, the ReLU function is very useful when training very deep neural networks with many layers, as seen from the 2012 AlexNet paper, in order to prevent vanishing gradients, where the network fails to update its weights from super small gradients
class Activation(Layer):
def __init__(self, activation, activation_prime):
self.activation = activation
self.activation_prime = activation_prime
def forward(self, input_val):
self.input = input_val
return self.activation(self.input)
def backward(self, output_gradient, optimizer):
return np.multiply(output_gradient, self.activation_prime(self.input))
def plot(self, x_min, x_max, points=25):
x = np.linspace(x_min, x_max, points)
y = self.activation(x)
y_prime = self.activation_prime(y)
fig, axes = plt.subplots(1, 2)
axes[0].plot(x, y)
axes[0].set_xlabel("X")
axes[0].set_ylabel("Y")
axes[0].set_title("F(X)")
axes[1].plot(x, y_prime)
axes[1].set_xlabel("X")
axes[1].set_ylabel("Y")
axes[1].set_title("F'(X)")
Tanh Activation Function#
We will use the Tanh activation function for our network:
\( \begin{align*} \sigma(z) &= \frac{e^z-e^{-z}}{e^z+e^{-z}} \end{align*} \)
Tanh Activation Function Gradient#
Since gradient descent relies on knowing the gradient of our activation function, let’s derivate the gradient of the tanh function.
\( \begin{align*} \sigma^\prime(z) &= \frac{\partial}{\partial z} [ \frac{e^z-e^{-z}}{e^z+e^{-z}} ] \\ &= \frac{(e^z+e^{-z})\frac{\partial}{\partial z}[e^z-e^{-z}] - (e^z-e^{-z})\frac{\partial}{\partial z}[e^z+e^{-z}]}{({e^z+e^{-z}})^2} \\ &= \frac{(e^z+e^{-z})(e^z+e^{-z}) - (e^z-e^{-z})(e^z-e^{-z})}{({e^z+e^{-z}})^2} \\ &= \frac{(e^z+e^{-z})^2 - (e^z-e^{-z})^2}{({e^z+e^{-z}})^2} \\ &= \frac{(e^z+e^{-z})^2}{({e^z+e^{-z}})^2} - \frac{(e^z-e^{-z})^2}{({e^z+e^{-z}})^2} \\ &= 1 - [\sigma(z)]^2 \end{align*} \)
Now that we have derived the gradient of the Tanh function, let’s create the class for the Tanh activation function.
class Tanh(Activation):
def __init__(self):
tanh = lambda x: np.tanh(x)
tanh_prime = lambda x: 1 - np.tanh(x) ** 2
super().__init__(tanh, tanh_prime)
Tanh().plot(-3, 3)
Optimizer#
Our optimization algorithm will be Gradient Descent, allowing us to determine how to update the parameters in the next iteration.
\( X_{n+1} = X_n - lr * \frac{\partial}{\partial X} f(X_n)\).
Let’s create a class that updates the weight matrix and the bias vector using the Gradient Descent equation.
class GradientDescentOptimizier:
def __init__(self, learning_rate):
self.learning_rate = learning_rate
def backward(self, weights, weights_gradient, bias, output_gradient):
weights -= self.learning_rate * weights_gradient
bias -= self.learning_rate * output_gradient
return weights, bias
Graphing Functions#
This Notebook will create many visualizations of the neural network during its training phase.
create_scatter_and_3d_plot() creates a 2 column graph for the Mean Squared Error graph on the left and a 3D graph on the right.
create_3d_and_3d_plot() creates a 2 column graph with two 3D graphs.
plot_3d_predictions() plots the neural network’s current preditions and the expected output of the neural network.
plot_layer_loss_landscape() plots how close one layer’s current weights are from the optimal weights by seeing how the total mean squared error changes if the weights were shifted a little.
import copy
def create_scatter_and_3d_plot():
fig, ax = plt.subplots(1, 3, figsize=(16 / 9.0 * 4, 4 * 1))
fig.suptitle("Neural Network Predictions")
ax[0].set_xlabel("Epoch", fontweight="normal")
ax[0].set_ylabel("Error", fontweight="normal")
ax[0].set_title("Mean Squared Error")
ax[1].axis("off")
ax[2].axis("off")
ax[2] = fig.add_subplot(1, 2, 2, projection="3d")
ax[2].set_xlabel("X")
ax[2].set_ylabel("Y")
ax[2].set_zlabel("Z")
ax[2].set_title("Function Approximation")
ax[2].view_init(20, -35)
ax[2].set_zlim(-1, 1)
ax[2].axis("equal")
camera = Camera(fig)
return ax[0], ax[2], camera
def create_3d_and_3d_plot():
fig, ax = plt.subplots(
1, 2, figsize=(16 / 9.0 * 4, 4 * 1), subplot_kw={"projection": "3d"}
)
fig.suptitle("Neural Network Loss Landscape")
ax[0].set_xlabel("W3_1")
ax[0].set_ylabel("W3_2")
ax[0].set_zlabel("MSE")
ax[0].set_title("Mean Squared Error")
ax[0].view_init(20, -35)
ax[0].set_zlim(-1, 1)
ax[0].axis("equal")
ax[1].set_xlabel("X")
ax[1].set_ylabel("Y")
ax[1].set_zlabel("Z")
ax[1].set_title("Function Approximation")
ax[1].view_init(20, -35)
ax[1].set_zlim(-1, 1)
ax[1].axis("equal")
camera = Camera(fig)
return ax[0], ax[1], camera
def plot_3d_predictions(ax, X, Y, Z, predictions, dims):
# Plot Neural Network Function Approximation
# Ground truth
ground_truth_legend = ax.plot_surface(
X, Y, Z, color="red", alpha=0.5, label="Ground Truth"
)
# Neural Network Predictions
predictions_legend = ax.scatter(
X,
Y,
predictions.reshape((dims, dims)),
color="blue",
alpha=0.2,
label="Prediction",
)
ax.plot_surface(
X,
Y,
predictions.reshape((dims, dims)),
color="blue",
alpha=0.3,
)
ax.legend(
(ground_truth_legend, predictions_legend),
("Ground Truth", "Predictions"),
loc="upper left",
)
def plot_layer_loss_landscape(
ax0,
network,
target_layer_idx,
features,
labels,
w1_min,
w1_max,
w2_min,
w2_max,
loss_dims,
):
# current target layer weights
target_layer_idx = target_layer_idx % len(network)
w1 = network[target_layer_idx].weights[0][0]
w2 = network[target_layer_idx].weights[0][1]
curr_error = 0
for x, y in zip(features, labels):
output = x
for layer in network:
output = layer.forward(output)
curr_error += mse(y, output)
curr_error /= labels.size
ax0.scatter([w1], [w2], [curr_error], color="red", alpha=0.4)
target_layer = copy.deepcopy(network[target_layer_idx])
w1_range = np.linspace(w1_min, w1_max, loss_dims)
w2_range = np.linspace(w2_min, w2_max, loss_dims)
w1_range, w2_range = np.meshgrid(w1_range, w2_range)
w_range = np.stack((w1_range.flatten(), w2_range.flatten()), axis=1)
error_range = np.array([])
for target_layer_weight in w_range:
target_layer_weight = target_layer_weight.reshape(1, 2)
target_layer.weights[0, :2] = target_layer_weight[0, :2]
error = 0
for x, y in zip(features, labels):
output = x
for layer_idx, layer in enumerate(network):
if layer_idx == target_layer_idx:
output = target_layer.forward(output)
else:
output = layer.forward(output)
error += mse(y, output)
error /= labels.size
error_range = np.append(error_range, error)
ax0.plot_surface(
w1_range,
w2_range,
error_range.reshape(loss_dims, loss_dims),
color="blue",
alpha=0.1,
)
def plot_mse_and_predictions(
ax0, ax1, idx, visible_mse, mse_idx, errors, X, Y, Z, predictions, dims
):
ax0.plot(
mse_idx[visible_mse][: idx + 1],
errors[visible_mse][: idx + 1],
color="red",
alpha=0.5,
)
plot_3d_predictions(ax1, X, Y, Z, predictions, dims)
def plot_loss_landscape_and_predictions(
ax0,
ax1,
network,
target_layer_idx,
features,
labels,
X,
Y,
Z,
predictions,
preds_dims,
w1_min=-5,
w1_max=5,
w2_min=-5,
w2_max=5,
loss_dims=20,
):
plot_3d_predictions(ax1, X, Y, Z, predictions, preds_dims)
plot_layer_loss_landscape(
ax0,
network,
target_layer_idx,
features,
labels,
w1_min,
w1_max,
w2_min,
w2_max,
loss_dims,
)
def show_epoch(epoch):
return (
epoch < 25
or (epoch < 25 and epoch % 2 == 0)
or (epoch <= 100 and epoch % 10 == 0)
or (epoch <= 500 and epoch % 25 == 0)
or (epoch <= 1000 and epoch % 50 == 0)
or epoch % 250 == 0
)
Training the model#
Let’s tie everything together to train the neural network. In each epoch, we will do the following:
Pass the training data into our model to get the model’s predictions
Calculate the loss from the model’s predictions and the expected value
Use the loss to run the optimizer to update the weights and biases
Call the visualization functions to visualize the training process
def fit(
network,
features,
labels,
X,
Y,
Z,
preds_dims,
epochs,
optimizer,
mse_plot_filename,
loss_landscape_plot_filename,
):
mse_idx = np.arange(1, epochs + 1)
errors = np.full(epochs, -1)
mse_ax, predictions_ax1, camera1 = create_scatter_and_3d_plot()
loss_landscape_ax, predictions_ax2, camera2 = create_3d_and_3d_plot()
network_len = len(network)
for idx in range(epochs):
error = 0
predictions = np.array([])
for x, y in zip(features, labels):
# Forward Propagation
output = x
for layer in network:
output = layer.forward(output)
predictions = np.append(predictions, output)
# Store Error
# no need to convert to numpy cpu, since both are tensors on device
error += mse(y, output)
# Backpropagation
grad = mse_prime(y, output)
for layer in reversed(network):
grad = layer.backward(grad, optimizer)
error /= len(X)
if show_epoch(idx):
print(f"epoch: {idx}, MSE: {error}")
# Plot MSE
errors[idx] = error
visible_mse = errors != -1
plot_mse_and_predictions(
mse_ax,
predictions_ax1,
idx,
visible_mse,
mse_idx,
errors,
X,
Y,
Z,
predictions,
preds_dims,
)
# plot the loss landscape of the second to last layer
# a 3D plot can be made because it's only 2 neurons
plot_loss_landscape_and_predictions(
loss_landscape_ax,
predictions_ax2,
network,
-2,
features,
labels,
X,
Y,
Z,
predictions,
preds_dims,
)
camera1.snap()
camera2.snap()
animation1 = camera1.animate()
animation1.save(mse_plot_filename, writer="pillow")
animation2 = camera2.animate()
animation2.save(loss_landscape_plot_filename, writer="pillow")
plt.show()
The model can be visualized with the following:

Our model consists of 3 layers with the Tanh() activation function after each layer.
Layer 1:
Input Dimensions: 2
Output Dimensions: 12
Layer 2:
Input Dimensions: 12
Output Dimensions: 2
Layer 3:
Input Dimensions: 2
Output Dimensions: 1
model = [Dense(2, 12), Tanh(), Dense(12, 2), Tanh(), Dense(2, 1), Tanh()]
Let’s train our model by passing our model and optimizer to our training method
epochs = 301
learning_rate = 0.005
optimizer = GradientDescentOptimizier(learning_rate)
mse_plot_filename = "neural_network.gif"
loss_landscape_plot_filename = "neural_network_loss_landscape.gif"
fit(
model,
features,
labels,
X,
Y,
Z,
dims,
epochs,
optimizer,
mse_plot_filename,
loss_landscape_plot_filename,
)
epoch: 0, MSE: 3.0033681391919607
epoch: 1, MSE: 2.861607692334731
epoch: 2, MSE: 2.825853287737221
epoch: 3, MSE: 2.817140865761648
epoch: 4, MSE: 2.814117393067351
epoch: 5, MSE: 2.8099737108937255
epoch: 6, MSE: 2.8023452137616185
epoch: 7, MSE: 2.7902417103220167
epoch: 8, MSE: 2.773063690822775
epoch: 9, MSE: 2.7502645093729323
epoch: 10, MSE: 2.721249705555072
epoch: 11, MSE: 2.685389899197139
epoch: 12, MSE: 2.642101512218608
epoch: 13, MSE: 2.590967280467696
epoch: 14, MSE: 2.531869391821693
epoch: 15, MSE: 2.4651083530357476
epoch: 16, MSE: 2.3914851384273637
epoch: 17, MSE: 2.3123291935128214
epoch: 18, MSE: 2.229456375261003
epoch: 19, MSE: 2.1450438173269872
epoch: 20, MSE: 2.061423945351905
epoch: 21, MSE: 1.9808288922004156
epoch: 22, MSE: 1.9051432156154482
epoch: 23, MSE: 1.8357262341875262
epoch: 24, MSE: 1.773340422003911
epoch: 25, MSE: 1.7181844787747815
epoch: 30, MSE: 1.5333665989875438
epoch: 40, MSE: 1.3570304555393908
epoch: 50, MSE: 1.2049292538302114
epoch: 60, MSE: 1.0420867215852363
epoch: 70, MSE: 0.8858079903889653
epoch: 75, MSE: 0.8162489390669957
epoch: 80, MSE: 0.7542965170101418
epoch: 90, MSE: 0.6530217909549149
epoch: 100, MSE: 0.5740174523365321
epoch: 125, MSE: 0.37933561711053515
epoch: 150, MSE: 0.24891189943713543
epoch: 175, MSE: 0.22012182602057515
epoch: 200, MSE: 0.20367177160551178
epoch: 225, MSE: 0.1908399435300111
epoch: 250, MSE: 0.1800010056452711
epoch: 275, MSE: 0.17054412799607388
epoch: 300, MSE: 0.16213374967702018
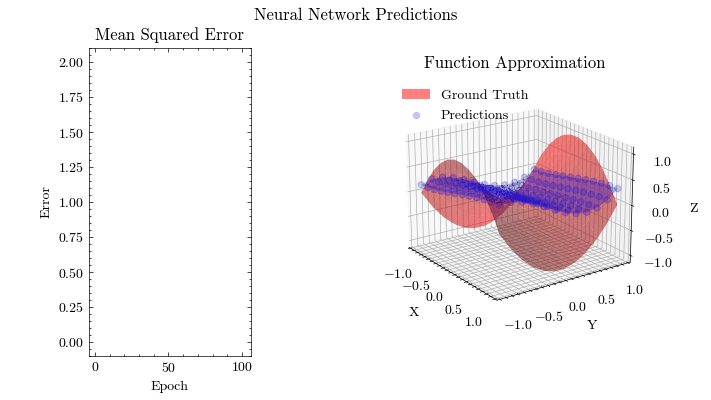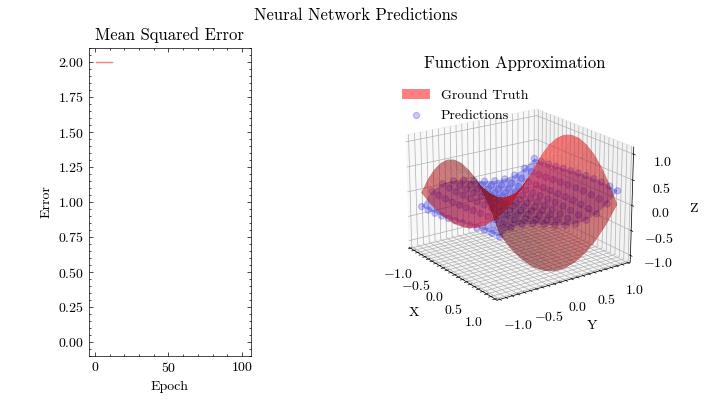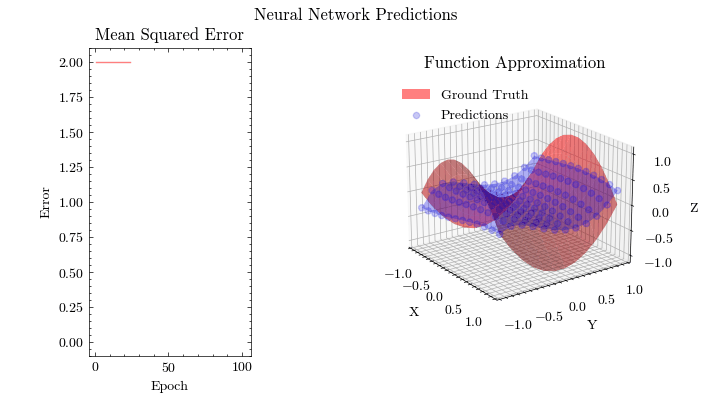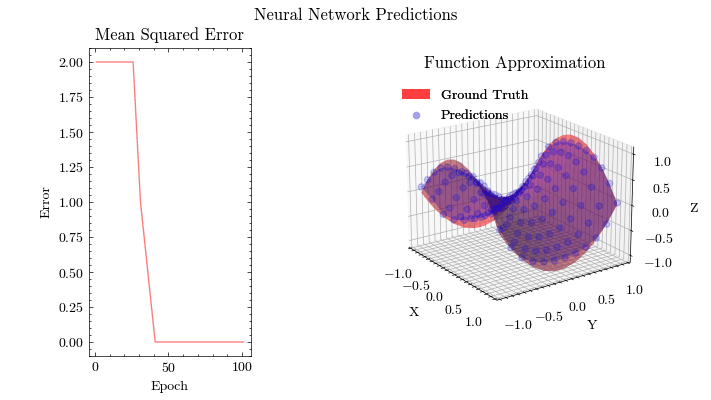
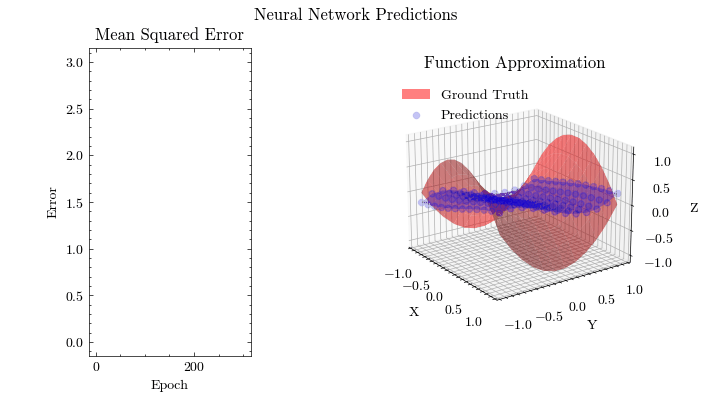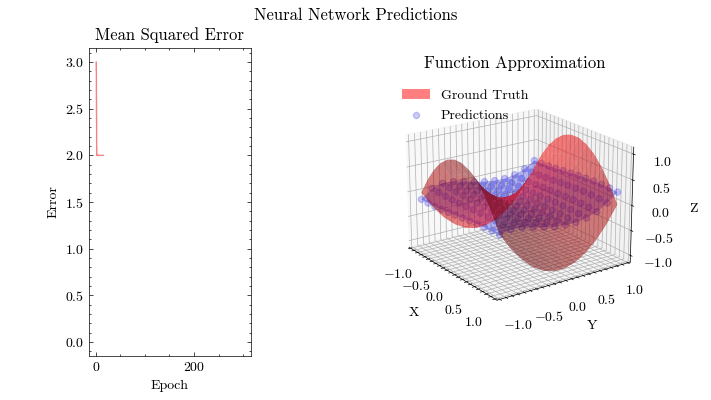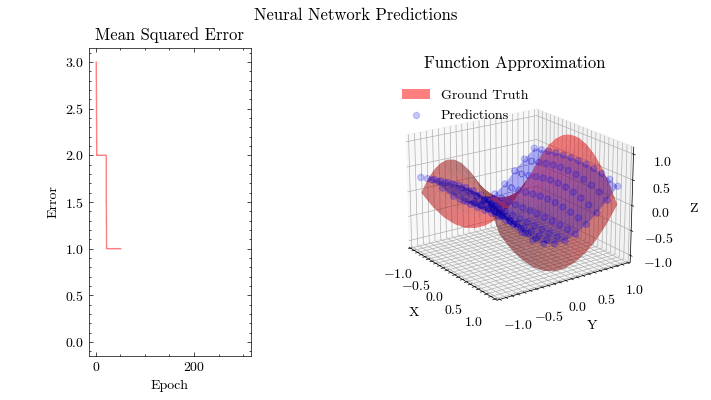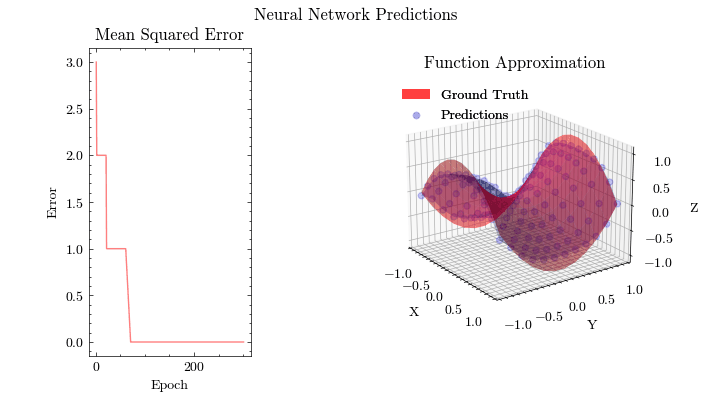
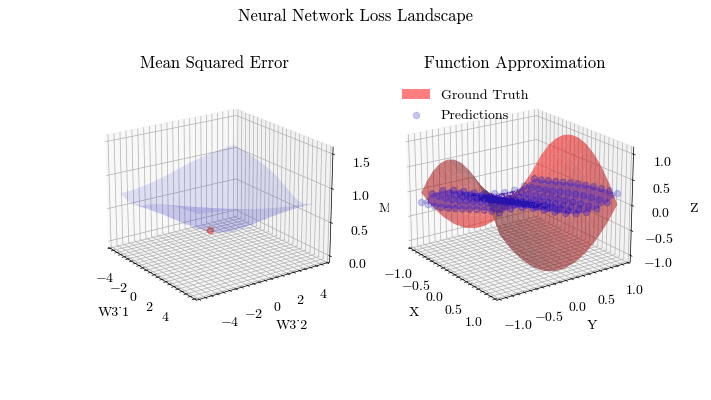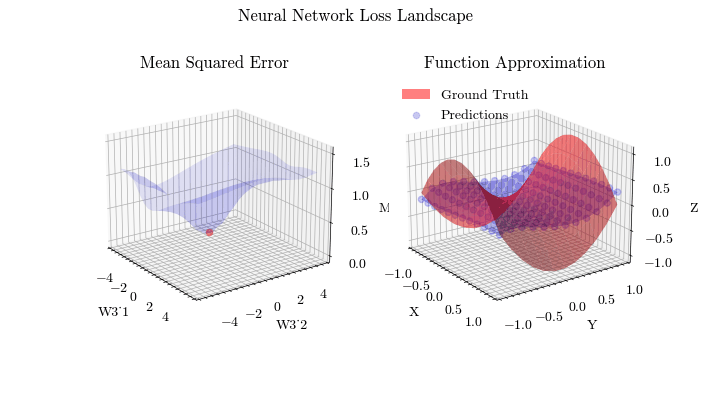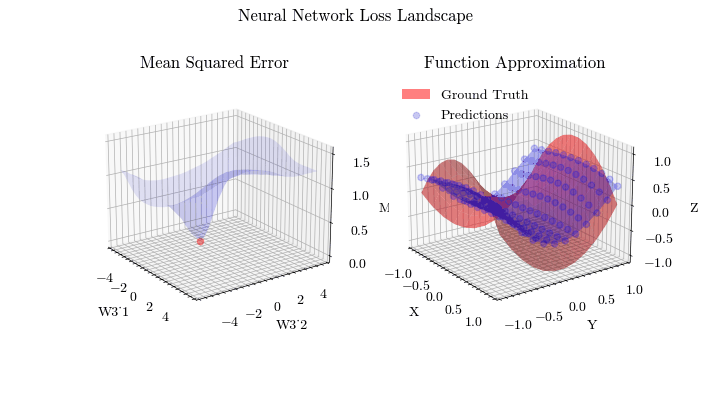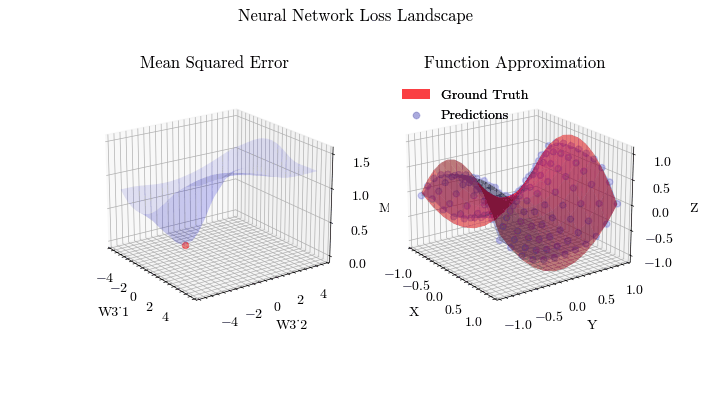
Output GIF#
In this visualization, we see the predictions fit the ground truth. The neural network was able to find the optimal parameters to fit this function. Now think about the applications of this. Given input data about people’s shopping habits, we can predict things to recommend to them. We can recommend a social media post to show them or a show to watch next. We can pass data to neural networks and it will uncover patterns from input data and find patterns.
Image(filename=mse_plot_filename)
The visualization below shows that backpropagation finds the optimal weights for the neural network. On the left graph, the red dot shows the current values of the weight matrix in the last layer of the neural network. The z axis is the mean squared error loss. If the weights weren’t at the current value, the loss wouldn’t be at a minima, meaning that backpropagation in fact does update the weights to the most optimal (local extrema) value and allows the network to converge.
Image(filename=loss_landscape_plot_filename)
Pytorch Implementation#
Machine Learning libraries, such as PyTorch, provide utilities to easily train and test neural networks on all types of optimized hardware. Now we will implement the same network in PyTorch.
import torch
import torch.nn as nn
import torch.nn.functional as F
torch.manual_seed(0)
<torch._C.Generator at 0xffff42d26ed0>
PyTorch nn.Module#
We represent our neural network by creating a subclass of the nn.Module PyTorch class that defines all of the layers in the network and how data flows through the network. The forward() method is the forward propagation of our neural forward. Notice that we don’t need to specify anything for the backpropagation process. PyTorch takes care of this for us using their auto-differentiation support. We just need to import an optimizer class, like the PyTorch provided gradient descent class, and pass in our model’s parameters. This is the beauty of using machine learning libraries.
class TorchNet(nn.Module):
def __init__(self):
super(TorchNet, self).__init__()
# define the layers
self.fc1 = nn.Linear(2, 14)
self.fc2 = nn.Linear(14, 2)
self.fc3 = nn.Linear(2, 1)
def forward(self, x):
# pass the result of the previous layer to the next layer
x = F.tanh(self.fc1(x))
x = F.tanh(self.fc2(x))
return F.tanh(self.fc3(x))
PyTorch Training#
Similar to our own implementation, let’s use our model to define our training process. In each epoch, we will do the following:
Pass the training data into our model to get the model’s predictions
Calculate the loss from the model’s predictions and the expected value
Use the loss to run the optimizer to update the weights and biases
Call the visualization functions to visualize the training process
def torch_fit(
model, features, labels, X, Y, Z, dims, epochs, optimizer, output_filename
):
mse_idx = np.arange(1, epochs + 1)
errors = np.full(epochs, -1)
mse_ax, predictions_ax1, camera1 = create_scatter_and_3d_plot()
loss_fn = nn.MSELoss()
for idx in range(epochs):
error = 0
predictions = np.array([])
for x, y in zip(features, labels):
# Forward Propagation
output = model(x)
output_np = output.detach().cpu().numpy()
predictions = np.append(predictions, output_np)
# Store Error
loss = loss_fn(output, y)
error += loss.detach().cpu().numpy()
# Backpropagation
optimizer.zero_grad()
loss.backward()
optimizer.step()
error /= len(X)
if show_epoch(idx):
print(f"epoch: {idx}, MSE: {error}")
# Plot MSE
errors[idx] = error
visible_mse = errors != -1
plot_mse_and_predictions(
mse_ax,
predictions_ax1,
idx,
visible_mse,
mse_idx,
errors,
X,
Y,
Z,
predictions,
dims,
)
camera1.snap()
animation1 = camera1.animate()
animation1.save(output_filename, writer="pillow")
plt.show()
Let’s call the training method and pass in the model, training data, and optimizer.
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
torch_model = TorchNet().to(device)
# the inputs and outputs for PyTorch must be tensors
features_tensor = torch.tensor(features, device=device, dtype=torch.float32).squeeze(-1)
labels_tensor = torch.tensor(labels, device=device, dtype=torch.float32).squeeze(-1)
epochs = 101
learning_rate = 0.005
optimizer = torch.optim.SGD(torch_model.parameters(), lr=learning_rate, momentum=0.0)
output_filename_pytorch = "neural_network_pytorch.gif"
torch_fit(
torch_model,
features_tensor,
labels_tensor,
X,
Y,
Z,
dims,
epochs,
optimizer,
output_filename_pytorch,
)
epoch: 0, MSE: 2.984565019607544
epoch: 1, MSE: 2.7075679302215576
epoch: 2, MSE: 2.6843478679656982
epoch: 3, MSE: 2.6776466369628906
epoch: 4, MSE: 2.681764841079712
epoch: 5, MSE: 2.690561294555664
epoch: 6, MSE: 2.700050115585327
epoch: 7, MSE: 2.7078511714935303
epoch: 8, MSE: 2.7127301692962646
epoch: 9, MSE: 2.7142038345336914
epoch: 10, MSE: 2.712219476699829
epoch: 11, MSE: 2.7069084644317627
epoch: 12, MSE: 2.698435068130493
epoch: 13, MSE: 2.686903715133667
epoch: 14, MSE: 2.672306776046753
epoch: 15, MSE: 2.654496908187866
epoch: 16, MSE: 2.6331663131713867
epoch: 17, MSE: 2.607834577560425
epoch: 18, MSE: 2.5778284072875977
epoch: 19, MSE: 2.5422799587249756
epoch: 20, MSE: 2.500131607055664
epoch: 21, MSE: 2.4501821994781494
epoch: 22, MSE: 2.3912034034729004
epoch: 23, MSE: 2.322108507156372
epoch: 24, MSE: 2.242227554321289
epoch: 25, MSE: 2.151536703109741
epoch: 30, MSE: 1.5780051946640015
epoch: 40, MSE: 0.4502718448638916
epoch: 50, MSE: 0.15558312833309174
epoch: 60, MSE: 0.10384205728769302
epoch: 70, MSE: 0.08925554901361465
epoch: 75, MSE: 0.08637455850839615
epoch: 80, MSE: 0.08450805395841599
epoch: 90, MSE: 0.0817754715681076
epoch: 100, MSE: 0.0793890580534935
Image(filename=output_filename_pytorch)